Section: New Results
Mixture models
Robust and collaborative extensions of Sliced Inverse Regression.
Participants : Stéphane Girard, Florence Forbes.
This research theme was supported by a LabEx PERSYVAL-Lab project-team grant.
Joint work with: A. Chiancone and J. Chanussot (Gipsa-lab and Grenoble-INP).
Sliced Inverse Regression (SIR) has been extensively used to reduce the dimension of the predictor space before performing regression. Recently it has been shown that this technique is, not surprisingly, sensitive to noise. Different approaches have thus been proposed to robustify SIR. In [16], we investigate the properties of an inverse problem proposed by R.D. Cook and we show that the framework can be extended to take into account a non-Gaussian noise. Generalized Student distributions are considered and all parameters are estimated via an EM algorithm. The algorithm is outlined and tested comparing the results with different approaches on simulated data. Results on a real dataset show the interest of this technique in presence of outliers.
For further improvement of SIR, in his PhD thesis work, Alessandro Chiancone studied the extension of the SIR method to different sub-populations. The idea is to assume that the dimension reduction subspace is not the same for different clusters of the data [17]. One of the difficulties is that standard Sliced Inverse Regression (SIR) has requirements on the distribution of the predictors that are hard to check since they depend on unobserved variables. It has been shown that, if the distribution of the predictors is elliptical, then these requirements are satisfied. In case of mixture models, the ellipticity is violated and in addition there is no assurance of a single underlying regression model among the different components. Our approach clusters the predictors space to force the condition to hold on each cluster and includes a merging technique to look for different underlying models in the data. A study on simulated data as well as two real applications are provided. It appears that SIR, unsurprisingly, is not able to deal with a mixture of Gaussians involving different underlying models whereas our approach is able to correctly investigate the mixture.
Structured mixture of linear mappings in high dimension
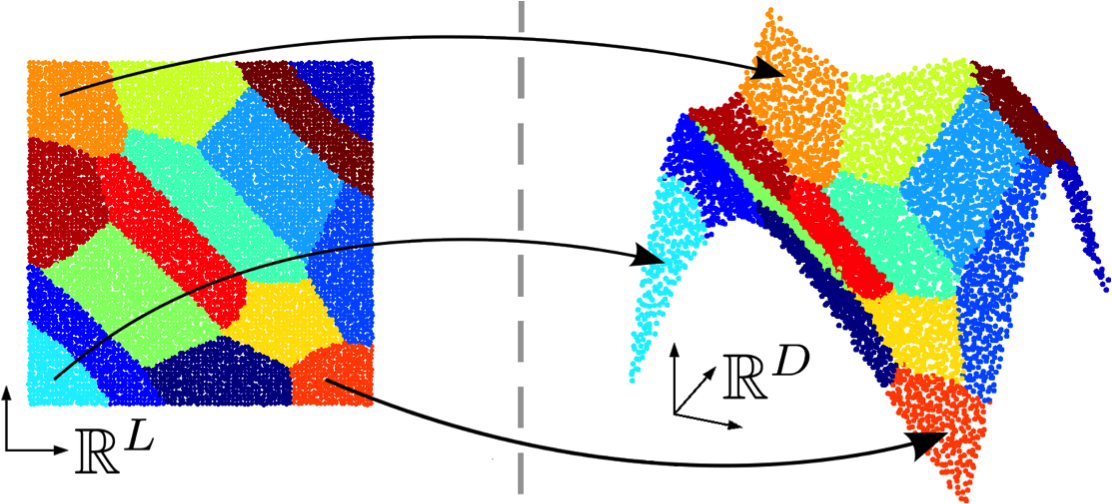
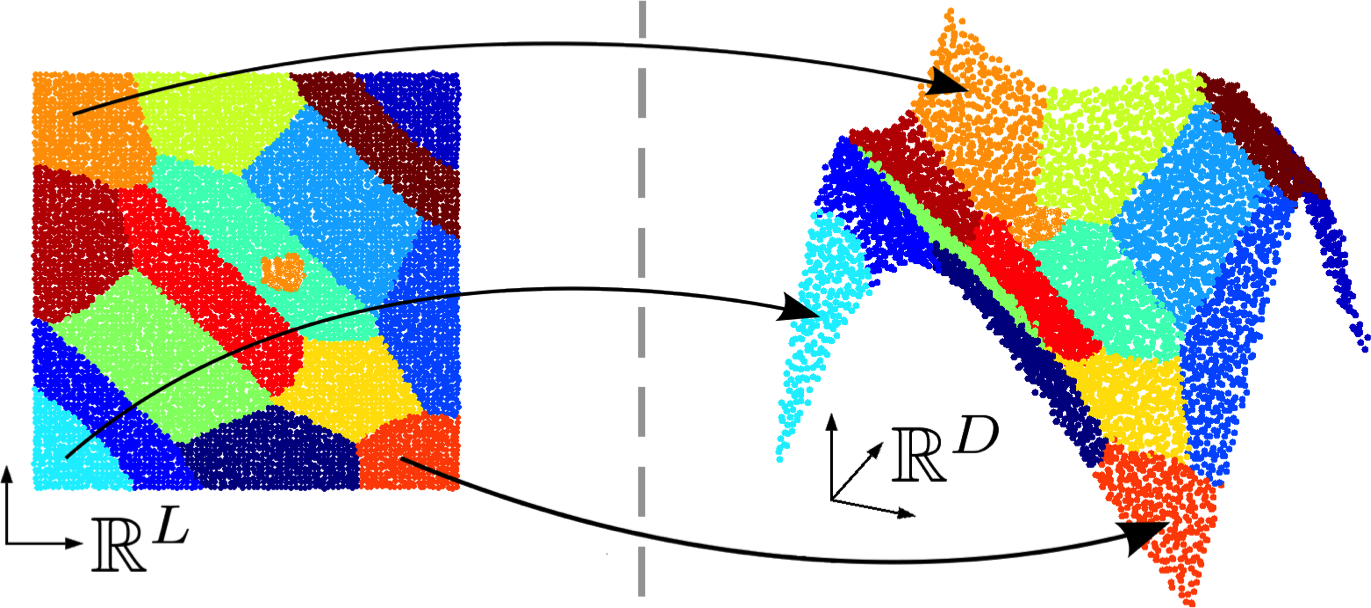
Participant : Florence Forbes.
Joint work with: Benjamin Lemasson from Grenoble Institute of Neuroscience, Naisyin Wang and Chun-Chen Tu from University of Michigan, Ann Arbor, USA.
Regression is a widely used statistical tool. A large number of applications consists of learning the association between responses and predictors. From such an association, different tasks, including prediction, can then be conducted. To go beyond simple linear models while maintaining tractability, non-linear mappings can be handled through exploration of local linearity. The non-linear relationship can be captured by a mixture of locally linear regression models as proposed in the so-called Gaussian Locally Linear Mapping (GLLiM) model [6] that assumes Gaussian noise models. GLLiM is based on a joint modeling of both the responses and covariates, observed or latent. This joint modeling allows for the use of an inverse regression strategy to handle the high dimensionality of the data. Mixtures are used to approximate non-linear associations. GLLiM groups data with similar linear association together. Within the same cluster, the association can be considered as locally linear, which can then be resolved under the classical linear regression setting (see Figure 2(a)). However, when the covariate dimension is much higher than the response dimension, GLLiM may result in erroneous clusters at the low dimension (eg Figure 2 (b)), leading to potentially inaccurate predictions. Specifically, when the clustering is conducted at a high joint dimension, the distance at low dimension between two members of the same cluster (component) could remain large. As a result, a mixture component might contain several sub-clusters and/or outliers, violating the model Gaussian assumption. This results in a model misspecification effect that can seriously impact prediction performance. A natural way to lessen this effect is to increase the number of components in the mixture making each linear mapping even more local. But this also increases the number of parameters to estimate and therefore requires to be done in a parsimonious manner to avoid over-parameterization. In this work, we propose a parsimonious approach which we refer to as Structured Mixture of Gaussian Locally Linear Mapping (SMoGLLiM) to solve the aforementioned problems. It follows a two-layer hierarchical clustering structure where local components are grouped into global components sharing the same high-dimensional noise covariance structure, which effectively reduces the number of parameters of the model. SMoGLLiM also includes a pruning algorithm for eliminating outliers as well as determining an appropriate number of clusters. Moreover, the number of clusters and training outliers determined by SMoGLLiM can be further used by GLLiM for improving prediction performance. As an extension, a subsetting and parallelization techniques are discussed for the efficiency concern. A preliminary version of this work was presented at the American Statistical Association Joint Statistical Meeting in Baltimore USA in July 2017, [35].
|
Dictionary-free MR fingerprinting parameter estimation via inverse regression
Participants : Florence Forbes, Fabien Boux, Julyan Arbel.
Joint work with: Emmanuel Barbier from Grenoble Institute of Neuroscience.
Magnetic resonance imaging (MRI) can map a wide range of tissue properties but is often limited to observe a single parameter at a time. In order to overcome this problem, Ma et al. introduced magnetic resonance fingerprinting (MRF), a procedure based on a dictionary of simulated couples of signals and parameters. Acquired signals called fingerprints are then matched to the closest signal in the dictionary in order to estimate parameters. This requires an exhaustive search in the dictionary, which even for moderately sized problems, becomes costly and possibly intractable . We propose an alternative approach to estimate more parameters at a time. Instead of an exhaustive search for every signal, we use the dictionary to learn the functional relationship between signals and parameters. This allows the direct estimation of parameters without the need of searching through the dictionary. We investigated the use of GLLiM that bypasses the problems associated with high-to-low regression. The experimental validation of our method is performed in the context of vascular fingerprinting The comparison between a standard grid search and the proposed approach suggest that MR Fingerprinting could benefit from a regression approach to limit dictionary size and fasten computation time. Preliminary tests and results have been submitted to ISMRM 2018, International Society for Magnetic Resonance in Medicine.
Semiparametric copula-based clustering
Participants : Florence Forbes, Gildas Mazo, Yaroslav Averyanov.
Modeling of distributions mixtures has rested on Gaussian distributions and/or a conditional independence hypothesis for a long time. Only recently have researchers begun to construct and study broader generic models without appealing to such hypotheses. Some of these extensions use copulas as a tool to build flexible models, as they permit to model the dependence and the marginal distributions separately. Recently [70], a semiparametric copula-based mixture model has been proposed to cluster continuous data. This semiparametric feature allows for more flexibility and reduces the modelling effort for the practitioner. Nonetheless, these advantages come at the cost of assuming that the clusters do not differ in scale. The aim of the internship of Y. Averyanov was to get rid of this assumption by building a nonparametric estimator which have to satisfy certain moment constraints. The performance of the estimator was tested on simulations and then embedded into an EM-like algorithm framework.
Fully automatic lesion localization and characterization: application to brain tumors using multiparametric quantitative MRI data
Participants : Florence Forbes, Alexis Arnaud.
Joint work with: Emmanuel Barbier, Nora Collomb and Benjamin Lemasson from Grenoble Institute of Neuroscience.
When analyzing brain tumors, two tasks are intrinsically linked, spatial localization and physiological characterization of the lesioned tissues. Automated data-driven solutions exist, based on image segmentation techniques or physiological parameters analysis, but for each task separately, the other being performed manually or with user tuning operations. In this work, the availability of quantitative magnetic resonance (MR) parameters is combined with advanced multivariate statistical tools to design a fully automated method that jointly performs both localization and characterization. Non trivial interactions between relevant physiological parameters are captured thanks to recent generalized Student distributions that provide a larger variety of distributional shapes compared to the more standard Gaussian distributions. Probabilistic mixtures of the former distributions are then considered to account for the different tissue types and potential heterogeneity of lesions. Discriminative multivariate features are extracted from this mixture modelling and turned into individual lesion signatures. The signatures are subsequently pooled together to build a statistical fingerprint model of the different lesion types that captures lesion characteristics while accounting for inter-subject variability. The potential of this generic procedure is demonstrated on a data set of 53 rats, with 36 rats bearing 4 different brain tumors, for which 5 quantitative MR parameters were acquired. This study has been submitted for publication [15].
Analyzing brain tumor tissue composition can then improve the handling of tumor growth and resistance to therapies. We showed on a 6 time point dataset of 8 rats that multiparametric MRI could be exploited via statistical clustering to quantify intra-lesional heterogeneity in space and time. More specifically, MRI can be used to map structural, eg diffusion, as well as functional, eg volume (BVf), vessel size (VSI), oxygen saturation of the tissue (StO2), characteristics. In previous work, these parameters have been analyzed to show the great potential of multiparametric MRI (mpMRI) to monitor combined radio- and chemo-therapies. However, to exploit all the information contained in mpMRI while preserving information about tumor heterogeneity, new methods need to be developed. We demonstrated the ability of clustering analysis applied to longitudinal mpMRI to summarize and quantify intra-lesional heterogeneity during tumor growth. This study showed the interest of a clustering analysis on mpMRI data to monitor the evolution of brain tumor heterogeneity. It highlighted the type of tissue that mostly contributes to tumor development and could be used to refine the evaluation of therapies and to improve tumor prognosis. This work has been presented at ISMRM 2017, International Society for Magnetic Resonance in Medicine [42].
Signature extraction in MR scans for de novo Parkinsonian patients
Participants : Florence Forbes, Veronica Munoz Ramirez, Julyan Arbel.
Joint work with: Michel Dojat from Grenoble Institute of Neuroscience.
This work is part of the cross-disciplinary project NeuroCoG. Parkinson's disease (PD) is characterized by the degeneration of dopaminergic neurons located in the substantia nigra pars compacta (SNc). This leads to well-known motor symptoms associated to Parkinson's disease, rigidity, akinesia and tremor. However, non-motor symptoms also appear. It is of primordial interest to understand these symptoms in order to optimize treatments and diagnose at an early stage the pathology's occurrence. The goal of the PhD work of Veronica Munoz Ramirez is the extraction of specific signatures from MR data of de novo PD patients. We investigated the possibility to use multivariate non-supervised clustering techniques as developed in the PhD thesis of Alexis Arnaud to cluster voxels taking into account interactions between various parameters.
Object-based classification from high resolution satellite image time series with Gaussian mean map kernels
Participant : Stéphane Girard.
Joint work with: C. Bouveyron (Univ. Paris 5), M. Fauvel and M. Lopes (ENSAT Toulouse)
In the PhD work of Charles Bouveyron [65], we proposed new Gaussian models of high dimensional data for classification purposes. We assume that the data live in several groups located in subspaces of lower dimensions. Two different strategies arise:
-
the introduction in the model of a dimension reduction constraint for each group
-
the use of parsimonious models obtained by imposing to different groups to share the same values of some parameters.
This modelling yielded a supervised classification method called High Dimensional Discriminant Analysis (HDDA)[4]. Some versions of this method have been tested on the supervised classification of objects in images. This approach has been adapted to the unsupervised classification framework, and the related method is named High Dimensional Data Clustering (HDDC)[3]. In the framework of Mailys Lopes PhD, our recent work [22], [23], consists in adapting this work to the classification of grassland management practices using satellite image time series with high spatial resolution. The study area is located in southern France where 52 parcels with three management types were selected. The spectral variability inside the grasslands was taken into account considering that the pixels signal can be modeled by a Gaussian distribution. To measure the similarity between two grasslands, a new kernel is proposed as a second contribution: the a-Gaussian mean kernel. It allows to weight the influence of the covariance matrix when comparing two Gaussian distributions. This kernel is introduced in Support Vector Machine for the supervised classification of grasslands from south-west France. A dense intra-annual multispectral time series of Formosat-2 satellite is used for the classification of grasslands management practices, while an inter-annual NDVI time series of Formosat-2 is used for permanent and temporary grasslands discrimination. Results are compared to other existing pixel- and object-based approaches in terms of classification accuracy and processing time. The proposed method shows to be a good compromise between processing speed and classification accuracy. It can adapt to the classification constraints and it encompasses several similarity measures known in the literature. It is appropriate for the classification of small and heterogeneous objects such as grasslands.